21 months ago by
United States
Hello,
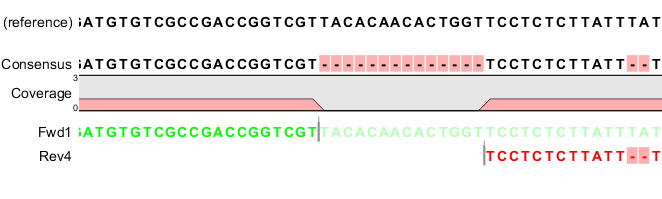
Coverage data can be calculated for BAM datasets using several tools. At http://usegalaxy.org, search in the tool panel with the keywords "bam coverage" or simply "coverage" (no quotes) to find these.
Assembly is different and doesn't always mean the same thing. Some tools generate an actual fasta consensus sequence and other describe the boundaries of the consensus sequence with respect to a reference genome/transcriptome. The type of input fastq and the final analysis goal make a difference in which tools are appropriate. For the primary choices, see the tool groups NGS: RNA Analysis, NGS: Du Novo, and NGS: Assembly. Other tools perform specific manipulations (example: BEDtools > GetFastaBed).
Even more tool choices are available in the Main Tool Shed for use in a local or cloud Galaxy.
If you want to share more about your analysis inputs and goals, we can help point you to tools that will work with that type of data. You can also review the suite of Galaxy tutorials here for guidance: https://new.galaxyproject.org/learn/
Thanks! Jen, Galaxy team