Question: How to count unique short sequences in FASTQ
0
yjoechen • 0 wrote:
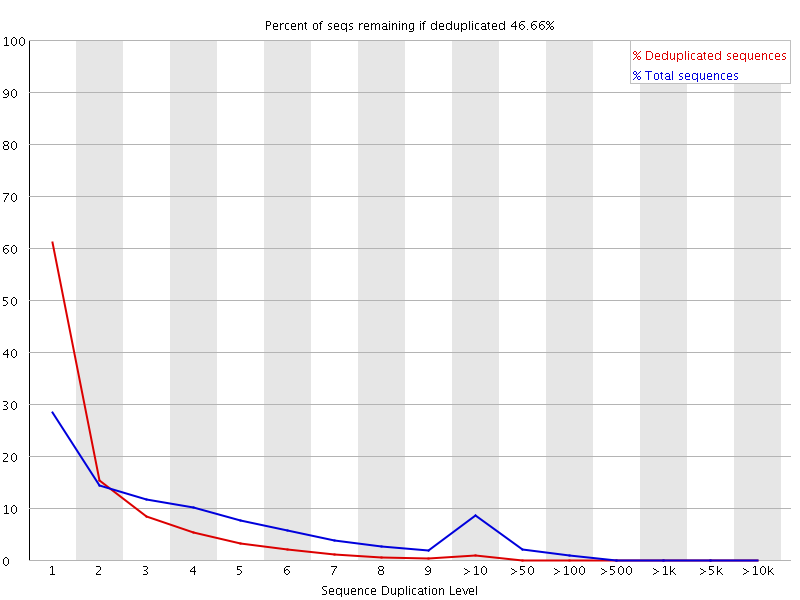
Hi, I wonder whether there is any tool that I could use to output the number of unique reads. I don't need any alignment to reference genome. Thanks!
ADD COMMENT
• link
•
modified 2.3 years ago
by
Devon Ryan • 1.9k
•
written
2.3 years ago by
yjoechen • 0